Single Cell & Spatial Multiomics
Headed by: Dr. Isabelle Scheyltjens & Dr. David Nittner
- Contact for untargeted / transcriptome-wide, sequencing-based single cell & spatial technologies
e-mail: isabelle.scheyltjens@kuleuven.be
location: O&N5, office 09.190
ORCID: 0000-0002-2260-2921
- Contact for targeted, imaging-based spatial platforms, immunohistochemical stainings, histological techniques:
David Nittner
ORCID: 0000-0002-2939-2453
For general questions/inquiries, please contact ccbspatialomics@ls.kuleuven.be
The tumor microenvironment is a unique and highly complex ecosystem in which tumor heterogeneity on the genetic, epigenetic, transcriptional and functional level importantly regulates cancer progression and therapy resistance. In the past decade our understanding of the interplay between cancer cells and non-cancerous cell types has greatly advanced with the emergence of single cell multiomic technologies. These allowed us to chart the heterogeneity of the tumor microenvironment, discover rare cell types and cell states, and predict the outcome of therapeutic strategies. Combined with low-plex technologies to visualize limited sets of markers within the tissue, our understanding of cancer biology has advanced considerably.
The presence of multiple functionally distinct cell types is linked to specific locations in space within a cancer- and/or patient-specific tumor microenvironment. Adding high-resolution spatial information to single cell technologies, and adding plexity to low-plex in situ visualization, is therefore crucial to fundamentally understand how tumor heterogeneity drives cancer progression, the interactions between cells within the tumor microenvironment, cancer cell clonality, and plasticity,... By consequence spatial technologies are required to improve therapy prediction and outcome.
Our expertise center is focused on the development and implementation of novel high-throughput single cell and high-resolution spatial technology platforms into the VIB Center for Cancer Biology. An important objective is to combine multiple levels of information by integrating data from multiomic technologies. As such we aim to optimize integration of high-plex proteomics with single cell and spatial genomics, epigenomics, transcriptomics, and metabolomics by collaboration with the CCB labs, the other CCB Expertise Centers and Core facilities.

Our Expertise
- Our expertise center focusses both on targeted and untargeted single cell and spatial -omics technologies, either sequencing-based, or imaging-based. Our scope of expertise ranges across:
- Single cell epigenomics
- Single cell transcriptomics
- Spatial epigenomics
- Spatial transcriptomics
- Spatial proteomics
- Spatial multiomics
- Inform the CCB research groups on a regular basis about novel interesting single cell or spatial (multi)-omics technologies
- Advise researchers from CCB labs based on an intake discussion
- Determine which technology is compatible with the research question and the available samples
- Assess the amount of optimization required for the technology and decide whether this falls within the scope of our Expertise Center, or whether the technology is available elsewhere within VIB
- Help with experimental design
- Advise on sample preparation
- Advise on funding possibilities
- Technology development, workflow optimizations, validation and implementation of new single cell or spatial (multi)-omics technologies:
- Technologies of interest for multiple CCB labs
- Early access technologies
- Recently published technologies
- Own technology development ideas
- New sample types or other substantial adaptations to existing technologies
- Perform comparative studies between different technology platforms
- Knowledge and SOP transfer to VIB CCB labs and collaborating VIB Core facilities
- Training of VIB CCB labs on workflows of multiomic technologies
- One of our main interests is the integration of spatial multiomic workflows and datasets preferentially on the same section, or on adjacent sections. This includes the integration of spatial epigenome data with spatial transcriptome, genome or proteome data. Through our collaboration with the Metabolism Expertise Center we also focus on the integration with spatial metabolomics.
- Our Expertise Center does not provide servicing, or bioinformatics analysis, however we aim to establish and closely collaborate with a bioinformatic community involved in spatial and multiomic integration pipelines creating an environment of easily accessible knowledge transfer to the research groups

Our Technologies
Single Cell Technologies
1. Fixed RNA Profiling
10x Genomics and other academic research groups have published multiple protocols in the past year which allow single cell RNA-Seq and combined protein profiling in fixed cells or tissues. By hybridizing a transcriptome-wide probe set, fragmented mRNA can be picked up and transcriptome-wide data can be obtained. In collaboration with the Single Cell Core we have access to all reagents and are currently optimizing fixed RNA profiling workflows for mouse and human samples of distinct tissue types. This platform allows multiplexing of samples, making it possible to collect and pool clinical samples prior to sequencing. As such, batch effects and costs per sample are lowered, while maintaining high-throughput scaled data-output. Depending on the tissue and cell-type of interest, we are currently testing the following workflows:
- Fixation of fresh or frozen cell- or nuclei-suspensions
- Fixation of fresh or frozen tissue prior to cell dissociation
- Isolation of cells or nuclei from FFPE archival tissue (10x Genomics and snPATHO-Seq (Vallejo et al., 2022, BioRxiv)
More information can be found on the following websites:
- https://www.10xgenomics.com/products/single-cell-gene-expression-flex
- https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-frp
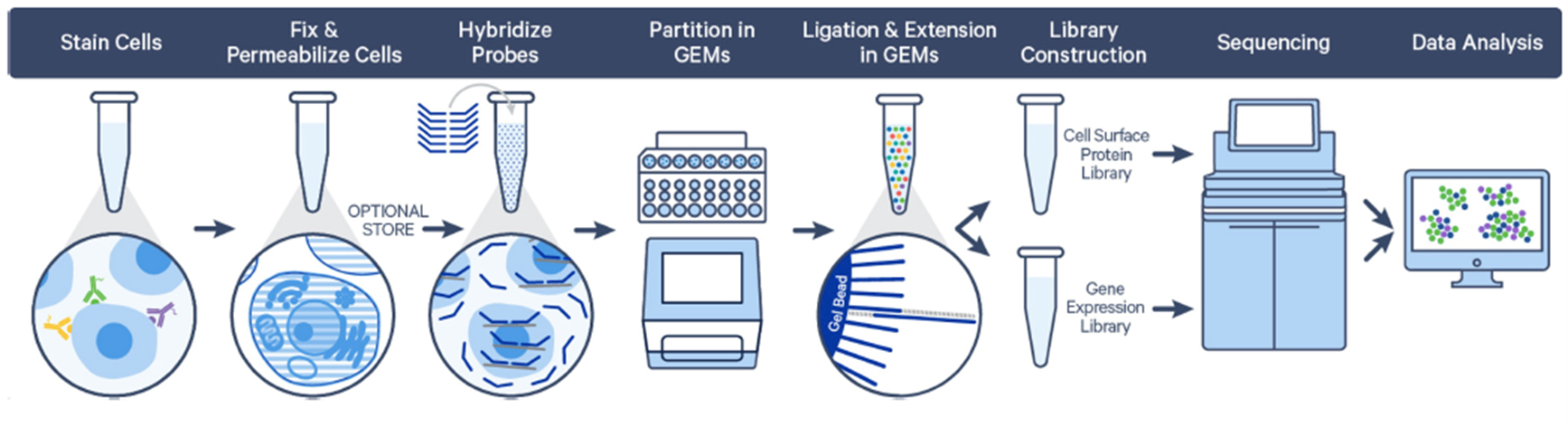
2. SPLiT-Seq (Parse Biosciences)
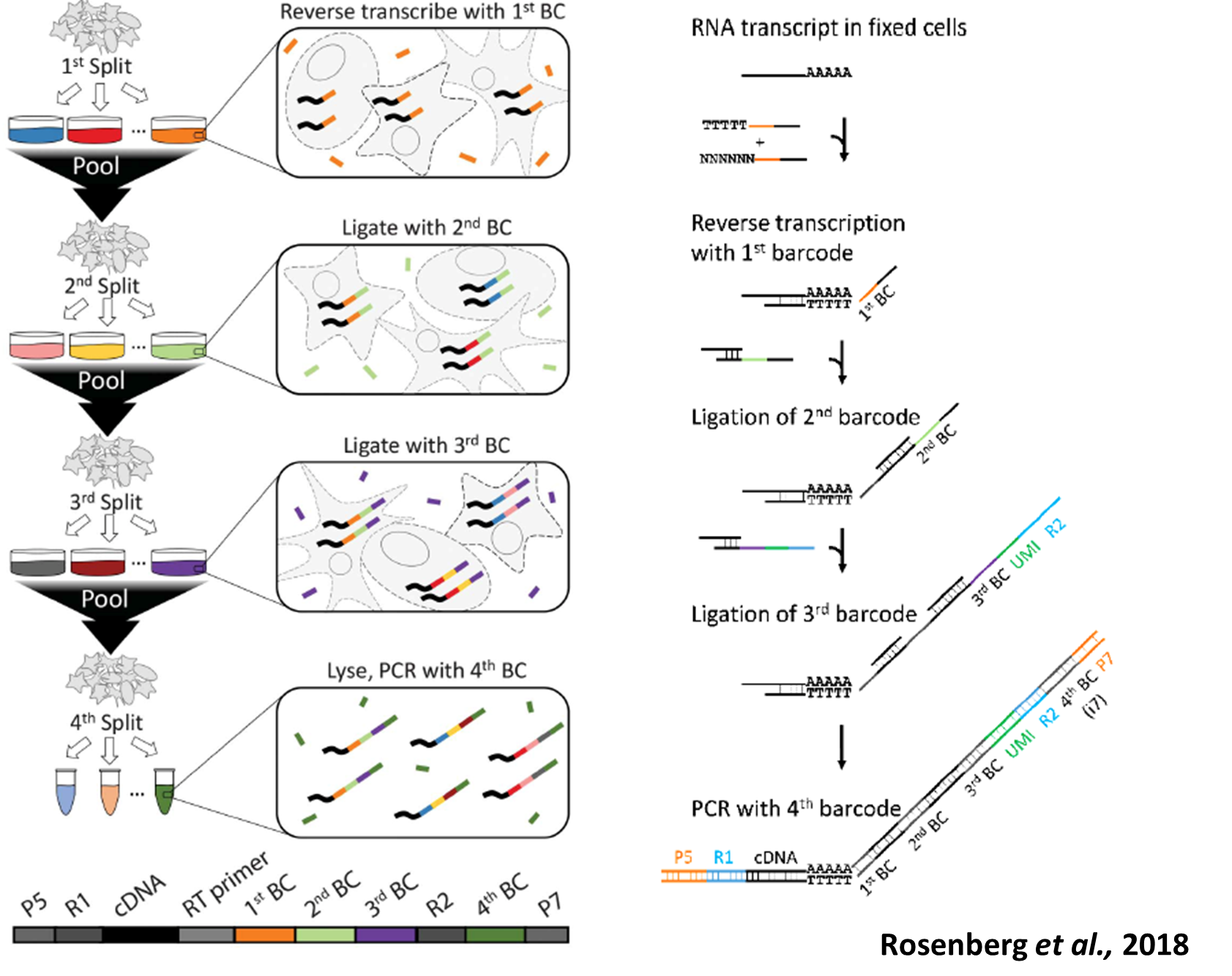
SPLiT-Seq (Split Pool Ligation-based Transcriptome sequencing) uses a combinatorial barcoding approach instead of droplet, microfluidics or well-based systems to capture single cells. This technology offers the advantages of having no cell type bias, low doublet count and lower cost while enabling high-throughput cell capturing. The technology works by multiple rounds of splitting and pooling fixed single cell or nuclei suspensions and ligating unique barcode sequences to the transcripts, generating unique barcode combinations for up to 1 million cells. Our current aim is to optimize these workflows to further decrease cell loss.
More information can be found on https://www.parsebiosciences.com and in Rosenberg et al.; 2018, Science.

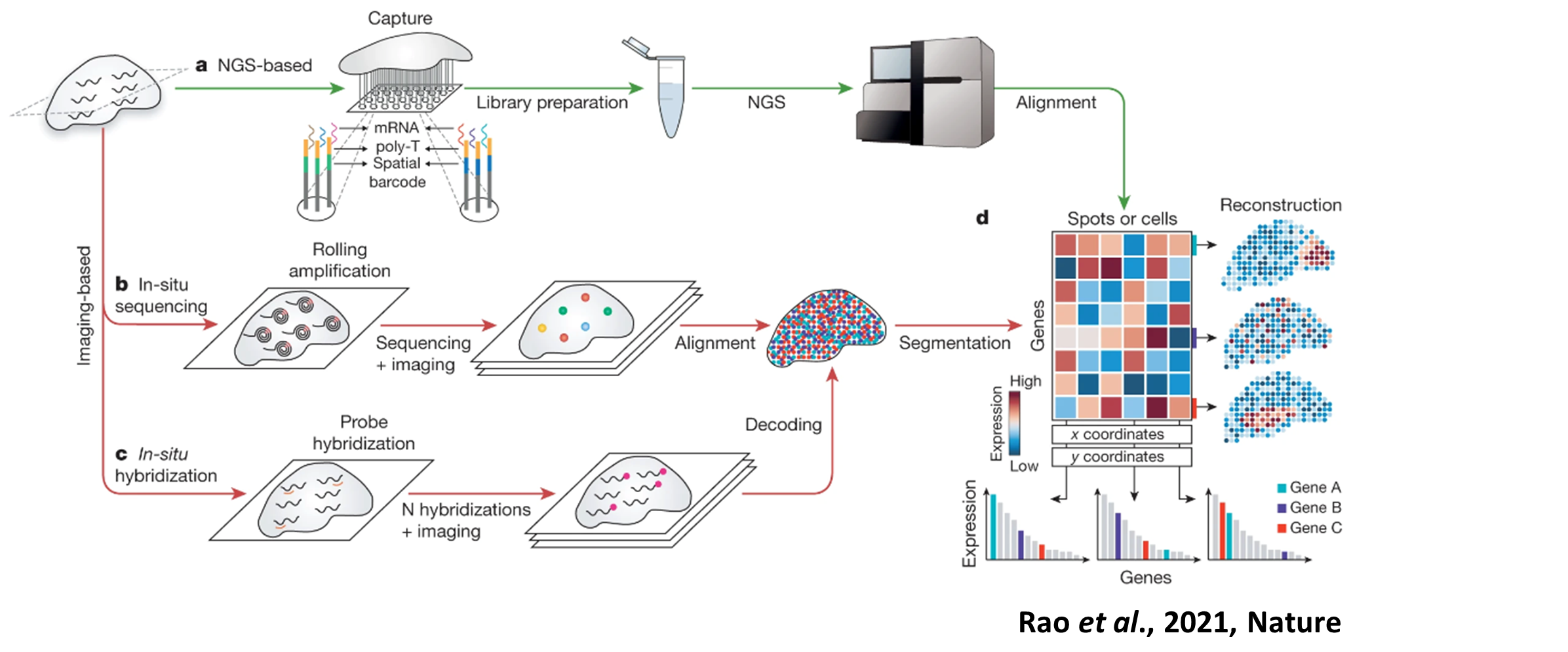
Spatial Technologies
Spatial transcriptomics technologies can broadly be divided into sequencing-based untargeted versus imaging-based targeted approaches. Each of these technologies scores differently in several key parameters, such as resolution, efficiency, sensitivity and field of view. Our expertise center keeps track of advances in these technologies and through discussions with the research groups, we can decide which trade-offs are most important in a particular project to choose the most fitting technology.
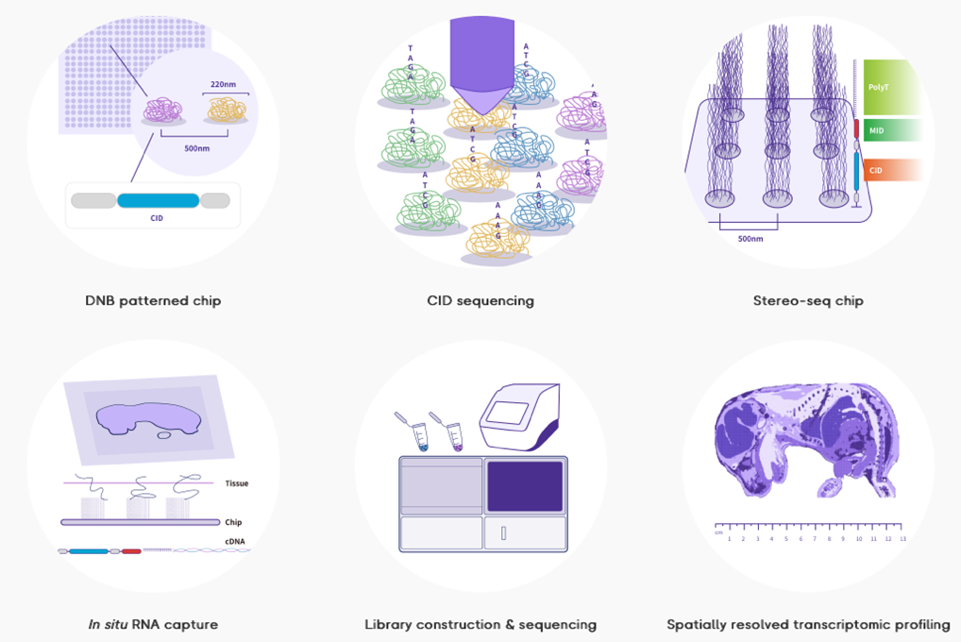
1. Stereo-Seq (STOmics; BGI research)
Stereo-Seq, or SpaTial Enhanced REsolution Omics-sequencing is one the emerging untargeted spatial transcriptomics technologies currently offering the highest (submicron) resolution and a large field of view (1 cm2 up to 42.25cm2) by using DNA-nanoball (DNB) technology. Within our Expertise Center we can currently provide full access, advise, and training to the entire Stereo-Seq workflow, from sample preparation, cryosectioning, mRNA capture and library preparation to sequencing. A DNB sequencer compatible with Stereo-Seq libraries (DNBSEQ-G400) is currently available at the Nucleomics Core. We closely monitor the status of novel developments, such as H&E co-staining protocols, FFPE tissue compatibility, spatial proteomics integration, and increased sensitivity, while also developing our own workflows for multiomic integration combined with Stereo-Seq.
More information can be found on https://www.stomics.tech.
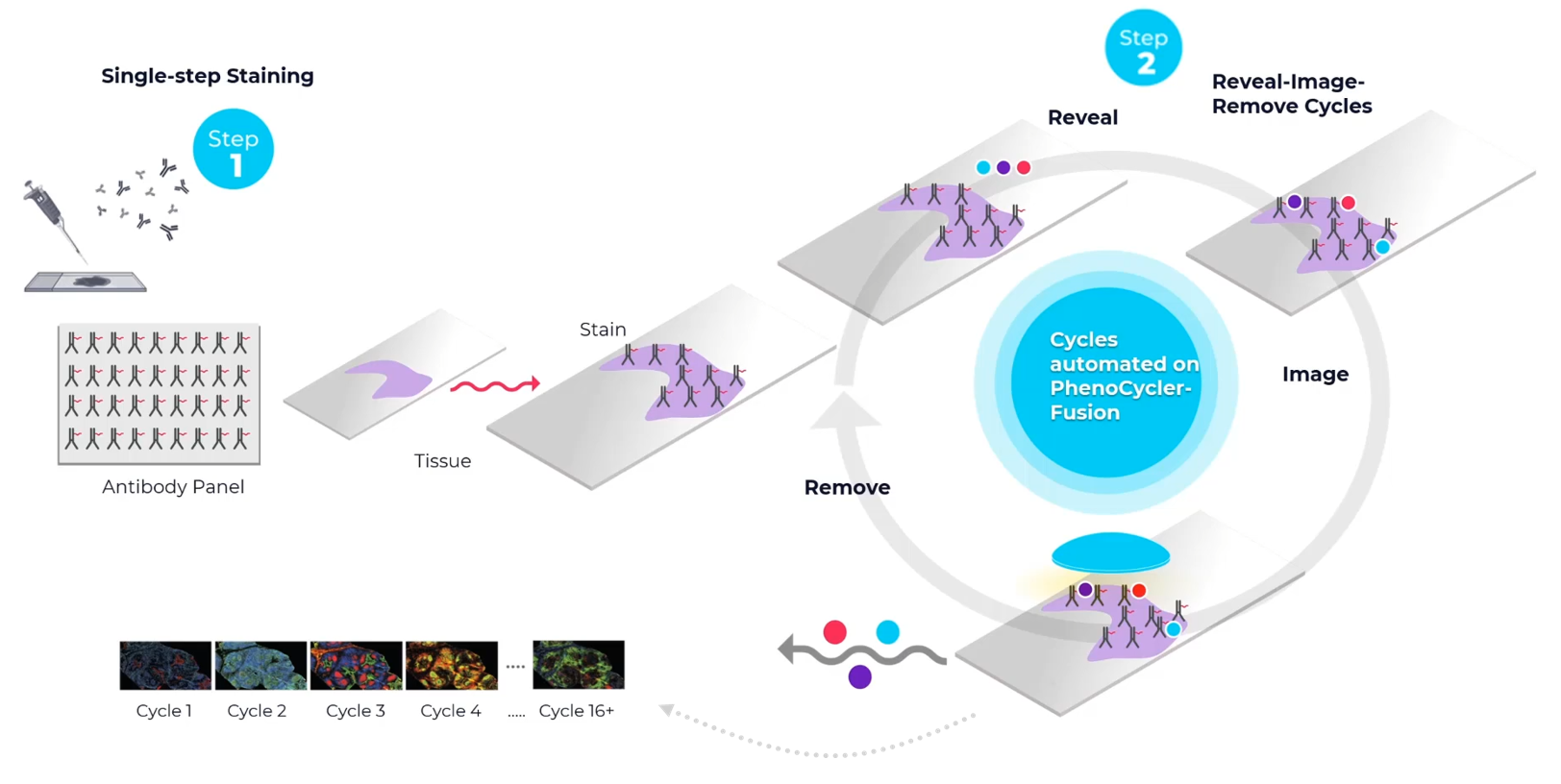
2. PhenoCycler Fusion (Akoya)
With the PhenoCycler Fusion system from Akoya it is possible to detect and visualize up to 100 protein targets on a single piece of tissue. This ultrahigh-plex staining method is combined with whole-slide imaging at single-cell resolution. Within our Expertise Center we can currently facilitate access to this platform, advise and train interested labs. This includes sample preparation, processing, staining, and basic analysis pipelines.
Find more information on https://www.akoyabio.com/phenocycler.
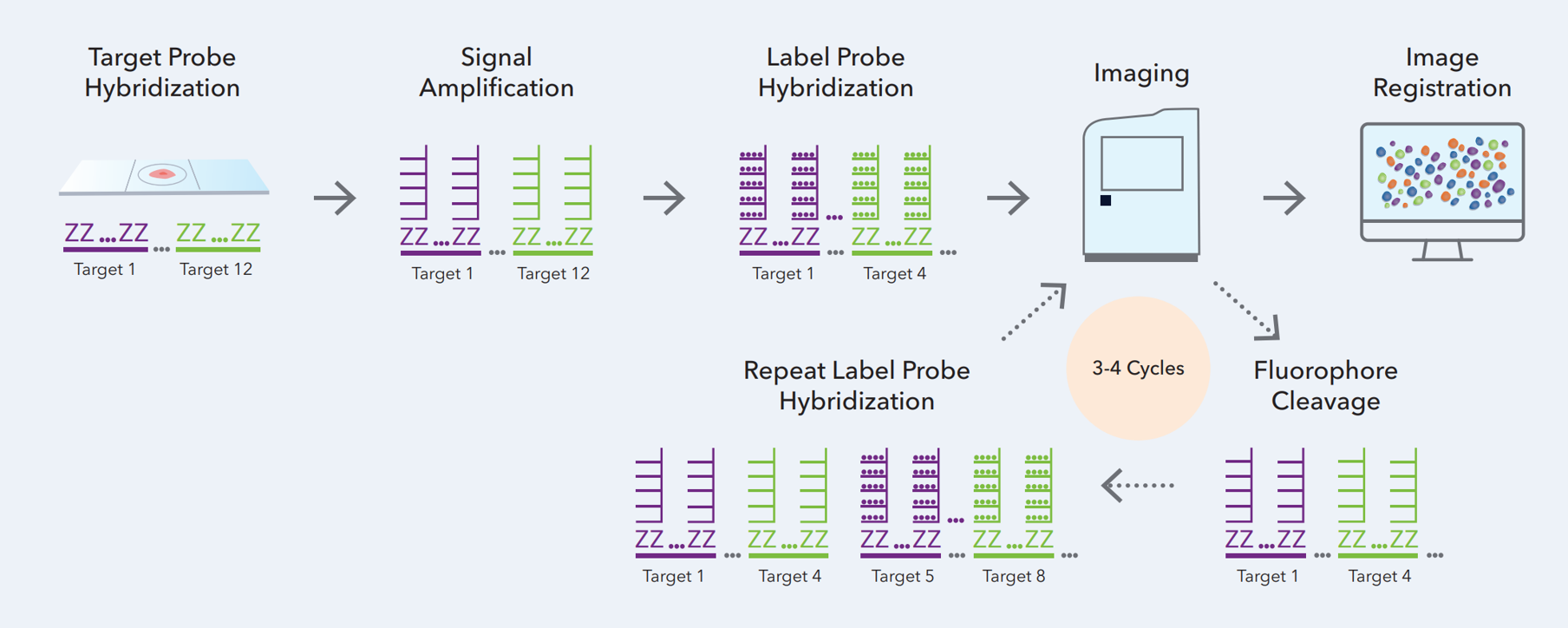
3. RNAscope HiPlex v2 (ACD)
The RNAscope HiPlex v2 assay is capable of multiplex fluorescent detection of up to 12 targets in fresh/fixed frozen and FFPE tissues. In this assay, after a series of highly effective and specific signal amplifications, the signal is detected iteratively, where up to four target genes are visualized in four distinct fluorescent channels. Similar to previous RNAscope assays, a single RNA transcript can be visualized as a punctate dot. The assay uniquely employs the cleavable versions of different fluorophores, and the iterative process is to be repeated until all 12 targets are detected. The fluorophore cleaving procedure is rapid with minimal to no effect on RNA and tissue morphology since it does not strip away the bound probe.
This new HiPlex v2 assay chemistry enables expansion of the multiplexing capability of the RNAscope HiPlex assay from 12 to 48-targets. The workflow contains a series of highly effective and specific signal amplifications, whereby running 4 cycles of 3 rounds each, allows for visualization of 4 targets by iterative detection after each round. Bringing it to a total number of 48 RNA targets.
Our Expertise Center can advise and train your lab in running the assay and - depending on your needs - try to combine the HiPlex v2 assay with other spacial technologies.
Find more information on: https://acdbio.com/rnascope-hiplex-assays.
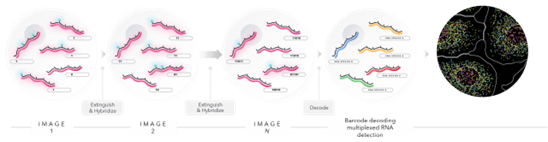
4. MERScope (VizGen)
MERFISH (Multiplexed Error-Robust Fluorescence in situ Hybridization) technology is built upon an RNA imaging-based technique called single molecule FISH. To perform smFISH, fluorescently tagged oligo probes label RNA transcripts of interest, and by directly counting the number of fluorescent RNA targets, you can determine each gene’s expression profile. MERFISH expands on single molecule FISH by adding combinatorial labeling, sequential rounds of imaging and error robust barcoding to greatly increase the multiplexing capacity and enable spatially resolved, single-cell gene expression profiling.
MERFISH implements a novel combinatorial barcoding scheme where each targeted gene in the custom designed panel is assigned a unique binary barcode. Sequential rounds of imaging are used to generate the barcode, a sequence of zeros and ones. This combinatorial barcoding scheme dramatically increases multiplexing capacity. (source: https://vizgen.com/technology/)
If you are interested in using the MERScope from VizGen our Expertise Center can help you to access and use it. MERScope uses the MERFISH technology to measure the copy number and spatial distribution of hundreds to tens of thousands of RNA species in individual cells at a subcellular resolution. One square centimeter can be profiled in a single run to detect custom multiplex panels from 100s of RNAs.
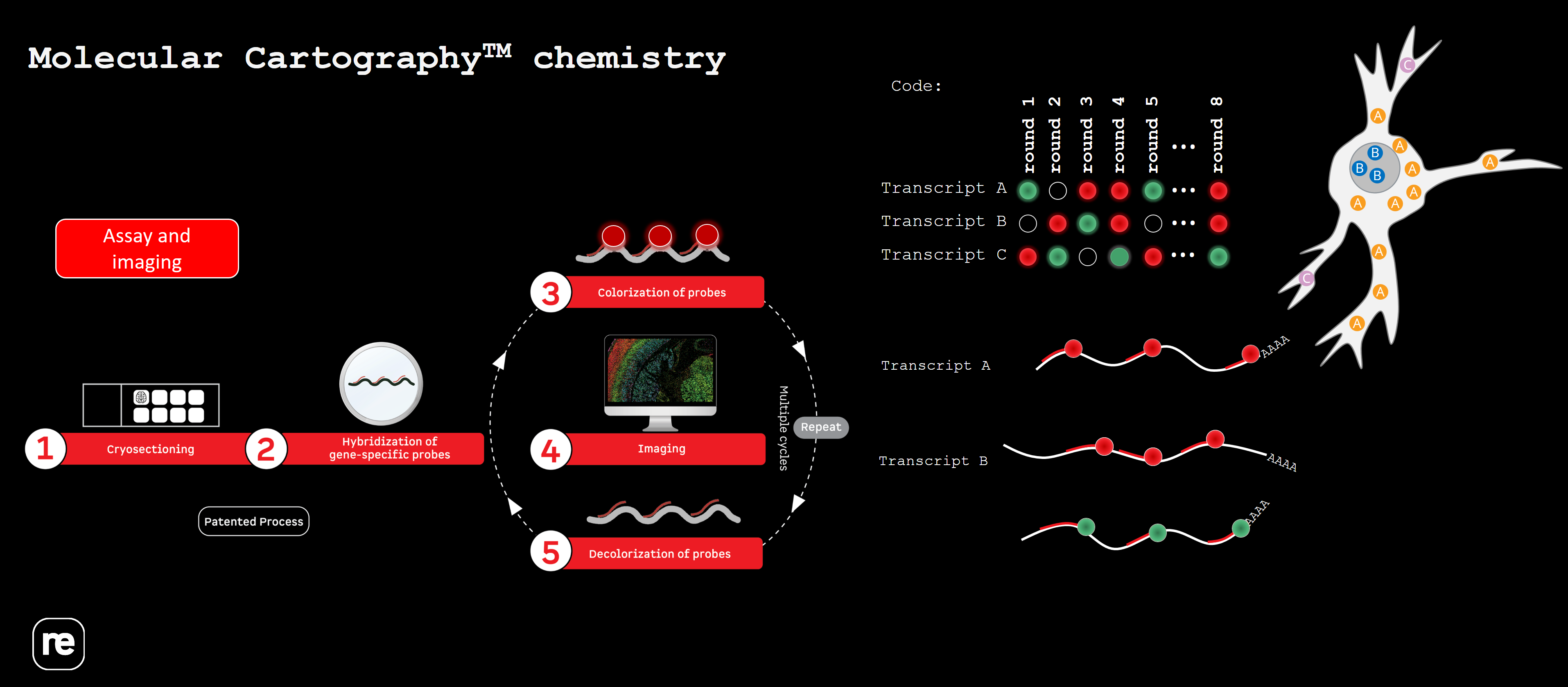
5. Molecular Cartography (Resolve Biosciences)
With Molecular Cartography from Resolve Biosciences it is possible to detect up to 100 RNA targets. These panels are fully customizable and are available for many species (human, mouse, drosophila,…). The technique of target detection relies on in situ barcoding in 8 rounds of imaging and has a subcellular resolution.
Our Expertise Center can help you to get access to the Resolve platform, advise and train your lab starting from sample preparation, sectioning, up to running the assay.
Find more information on: https://resolvebiosciences.com/.